Development¶
The source code is on GitHub
Code structure¶
pyKNEEr is written in a modular way to favor modifications and extensions. There are three levels of files associated with each step of the workflow:
Jupyter notebooks (
.ipynb) representing both a user-interface and a record of the method and results obtained, to be attached to publicationsPython scripts for notebooks (
for_nb.py) containing functions called by the notebook and used to process images and visualize outputsBasic python scripts (
.py) containing core functions and classes
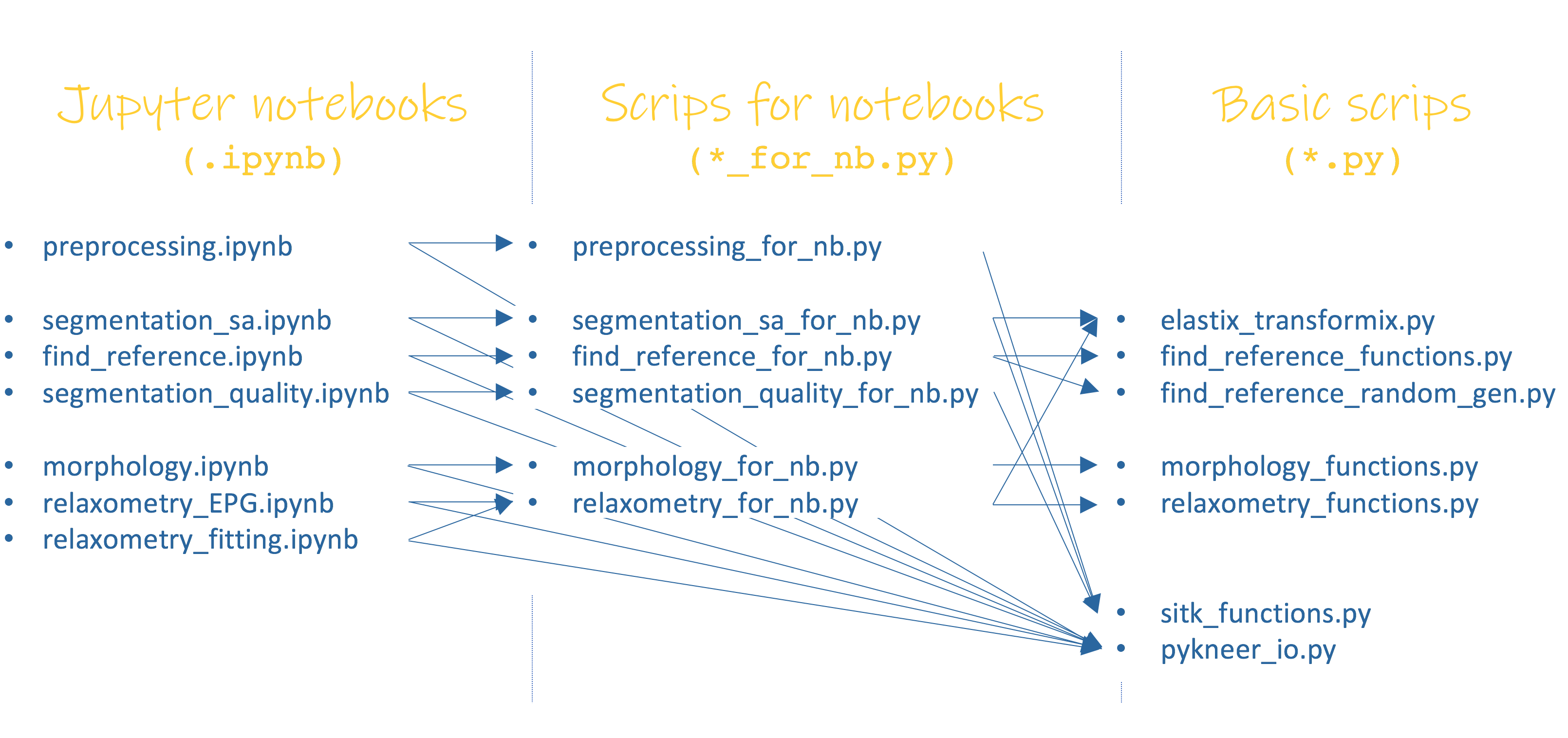
The figure above shows the code structure. For example, to compute morphology of femoral knee cartilage, the scheme is:
morphology.ipynb: Jupyter notebook used to run computations and visualize results. It uses functions from:morphology_for_nb.py: python module that contains functions to calculate knee cartilage morphology and visualize results. It uses functions from:morphology_functions.py: python module that contains the actual algorithms to compute cartilage thickness and volume
Additional packages¶
pyKNEEr uses:
Elastix 4.8 for single-atlas segmentation
Cylinder fitting to flatten cartilage for visualization
Variable names¶
When reading the .txt file at the beginning of each notebook, pyKNEEr creates a dictionary (i.e. a struct) called image_data.
In image_data, each cell corresponds to an image and contains an array of strings representing paths, input and output file names, and other information that will be used throughout the whole notebook.
For example, for the demo images the content of the first cell of image_data in the preprocessing.ipynb notebook can be printed out using the commands:
image_data = io.load_image_data_preprocessing(input_file_name)
image_data[0] # 0 is the ID of the first image
obtaining the following output:
Folders |
Example |
|---|---|
original_folder |
- /.original |
preprocessed_folder |
/.preprocessed/ |
image_folder_file_name |
01/DESS/01 |
File Names |
Example |
original_file_name |
./preprocessed/01_DESS_01_orig.mha |
preprocessed_file_name |
./preprocessed/01_DESS_01_prep.mha |
info_file_name |
./preprocessed/01_DESS_01_orig.txt |
temp_file_name |
./preprocessed/01_DESS_01_temp.mha |
Other Information |
Example |
image_name_root |
subject1_DESS |
laterality |
right |
