Segmentation¶
pyKNEEr computes atlas-based segmentation, which is based on registration, using elastix [1]. In registration there are a reference (or target) image and a moving (or floating) image, which is warped to the reference image. The reference image is already segmented, whereas the moving image has to be segmented.
In pyKNEEr, atlas-based segmentation has three steps:
The moving image is registered to the reference image though transformations
The transformations are inverted
The inverted transformations are applied to the reference mask to obtain the moving mask
These steps are applied:
To the femur to align the moving image to the reference image, and guide femoral cartilage segmentation
To the femoral cartilage to obtain the segmented image
Important
The current method does not take into account segmentation accuracy at the bone-cartilage interface. The femur alignment is used to guide cartilage segmentation, and femur segmentation is a byproduct of the workflow.
In pyKNEEr, there are three segmentation modalities:
New subject: Segmentation of single images, baseline images in longitudinal studies, or high-resolution images in multimodal acquisitions
Multimodal: Segmentation of images acquired with different protocols, where the highest resolution image has already been segmented as new subject
Longitudinal: Segmentation of followup images, where the baseline image has already been segmented as new subject
For the execution, the differences among the three modalities are:
The reference image
The structure of the input file
The variable
modalityinsegmentation_sa.ipynb
New subject segmentation¶
Reference image¶
The reference image is the same for all the images to be segmented
In the folder reference, the reference image is in the folder newsubject, which contains:
reference.mha: Reference image for the atlas-based segmentationreference_f.mha: Femur mask of the reference imagereference_fc.mha: Femoral cartilage mask of the reference image.
This reference was found with a convergence study on 19 segmented images
Tip
When using your own data:
You can use the same reference image contained in the demo dataset: copy the files
reference.mha,reference_f.mha, andreference_fc.mhato yourreference/newsubjectfolderYou can use a different reference image: copy your new reference image, femur mask, and femoral cartilage mask to the
reference/newsubjectfolder, making sure you rename the files asreference.mha,reference_f.mha, andreference_fc.mhaIf you want to find a reference image from an already segmented dataset, you can run pyKNEEr convergence study
Input: Image list¶
For the demo images, the input file is image_list_newsubject.txt, which contains:
[1] ./reference/newsubject
[2] ./preprocessed/
[3] r reference.mha
[4] m 01_DESS_01_prep.mha
where:
Line 1: Reference folder, containing the reference image and its masks
Line 2: Preprocessed file folder, containing the preprocessed files
Line 3: Reference image, indicated as
rLine 4: Moving images, indicated as
m
Tip
When using your own data:
Customize
image_list_newsubject.txtwith the paths and the names of your imagesThere is no limit to the number of moving
mimages
Executing segmentation_sa_ns.ipynb¶
To segment the data:
Launch Jupyter notebook
In File Browser, navigate to
segmentation_sa_ns.ipynb, open it, and:Customize the input variable
n_of_cores(How do I choose the number of cores?)Notice that variable
modalityis set tonewsubjectFollow the instructions in the notebook
Save your notebook at the end of the process
Output: Segmented images¶
The masks are in the folder segmented. For each subjects, the outputs are:
*_prep_fc.mha(e.g.01_DESS_01_prep_fc.mha): Binary mask of the femoral cartilage*_prep_f.mha(e.g.01_DESS_01_prep_f.mha): Binary mask of the femur, a byproduct of the registration
Note
Intermediate registration steps are saved in the folder
registeredIf you are not interested in analysis from deformations, you can delete the folder after your computations
If you want to compute further analysis, the folders
registered/subject_namecontain:fc_spline.mha(intersubject and longitudinal segmentation) orf_rigig.mha(multimodal segmentation), which contain the moving image warped to the reference. They can be used for analysis such as voxel-based relaxometryTransformParameters.xxx.txt, which contain transformation values. They can be used for PCA or other analysis. For their use, we forward to the elastix manual
Visualization: Superimposing cartilage mask onto the MR image¶
For a qualitative check, for each subject we visualize three 2D slices of the intensity image (*_prep.mha) overlapped by the corresponding slices of the cartilage mask (*_prep_fc.mha), similarly to this figure:
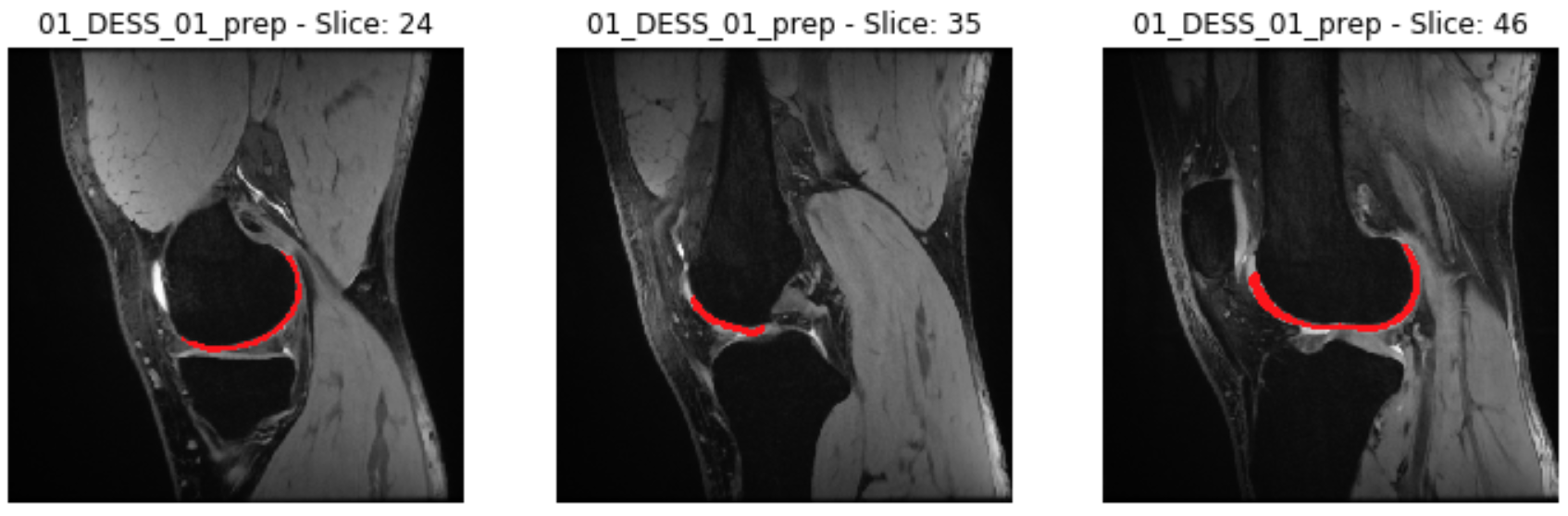
For a 3D check, consider using a medical image software such as ITK-SNAP, which allows visualizing the overlap of an image and its mask
Multimodal segmentation¶
Reference image¶
For each acquisition at lower resolution (e.g. CubeQuant), the reference image is a high-resolution image of the same subject (e.g. DESS), which must have been previously segmented as a newsubject.
In the folder reference, create the folder multimodal, and copy:
The high-resolution image:
01_DESS_01_prep.mhafrom the folderpreprocessedThe high-resolution femur mask:
01_DESS_01_prep_f.mhafrom the foldersegmentedThe high-resolution femoral cartilage mask:
01_DESS_01_prep_fc.mhafrom the foldersegmented
This step will be simplified in future versions of pyKNEEr
Tip
When using your own data:
In the folder
referencecreate the foldermultimodalCopy the high-resolution images to be used as a references, together with their femur mask and femoral cartilage mask
Input: Image list¶
For the demo images, the input file is image_list_multimodal.txt, which contains:
[1] ./reference/multimodal
[2] ./preprocessed/
[3] r 01_DESS_01_prep.mha
[4] m 01_cubeQuant_01_prep.mha
where:
Line 1: Reference folder, containing the baseline images used as reference
Line 2: Preprocessed file folder, containing the preprocessed images
Line 3: Reference (high res) image, indicated as
rLine 4: Moving (low res) image, indicated as
m
Tip
When using your own data:
Customize
image_list_multimodal.txtwith the paths and the names of your imagesIn case of several images to segment, write high-resolution images
rand low-resolution imagesmin a coupled manner:[1] ./reference/longitudinal [2] ./preprocessed/ [3] r subject1_HRes_prep.mha [4] m subject1_LRes_prep.mha [5] r subject2_HRes_prep.mha [6] m subject2_LRes_prep.mha [7] r subject3_HRes_prep.mha [8] m subject3_LRes_prep.mha [9] etc.
Execution, Output, and Visualization¶
Execution:
To segment the data, apply the instructions above to the notebook
segmentation_sa_mm.ipynb. Note that the variablemodalityis set tomultimodal
Output and visualization:
Longitudinal¶
For this segmentation modality, we do not provide a demo example but only instructions as it is very similar to multimodal segmentation
Reference image¶
For each followup image, the reference image is the corresponding baseline image, which must have been previously segmented as a new subject
In the folder reference, create the folder longitudinal, and for each image copy:
The baseline image:
BL_prep.mhaThe baseline femur mask:
BL_prep_f.mhaThe baseline femoral cartilage mask:
BL_prep_fc.mha
This step will be simplified in future versions of pyKNEEr.
Input: Image list¶
Create the file image_list_longitudinal.txt, which will contain:
[1] ./reference/longitudinal
[2] ./preprocessed/
[3] r subject1_BL_prep.mha
[4] m subject1_FU_prep.mha
[5] r subject2_BL_prep.mha
[6] m subject2_FU_prep.mha
[7] r subject3_BL_prep.mha
[8] m subject3_FU_prep.mha
where:
Line 1: Reference folder, containing the the baseline images used as reference
Line 2: Preprocessed file folder, containing the preprocessed files of the corresponding followup images
Odd lines from 3 to 7: Reference (baseline) images, indicated as
rEven lines 4 to 8: Moving (followup) images, indicated as
m
Execution, Output, and Visualization¶
Execution:
To segment the data, apply the instructions above. Set the variable
modalitytolongitudinal
Output and visualization:
Segmentation Plus¶
pyKNEEr includes notebooks to find the reference image and evaluate segmentation quality
These two steps are not included in the demo for sake of simplicity
Finding reference image¶
In this convergence study, the new reference is the image of the dataset whose vector field is the closest to the average of the vector fields of the dataset. The study runs until convergence or for a fixed amount of iterations
Note
To run this convergence study all the images of the dataset must already have a femur mask }
The Jupyter notebook to find a reference image is find_reference.ipynb
Picking random seeds¶
To determine the image that you are going to use as reference, we recommend a random generator function with a fixed seed to make the reference selection reproducible. The code is here.
You can run several convergence study in parallel to confirm you find the same reference image independently from the starting seed
Input: Image list¶
Data required are MR images of the knee that have segmented femurs because:
the registration is guided by the femur mask
the average vector field is calculated in the femur mask
the comparison between the average vector field and each image vector field is performed in the femur mask
In you data folder, create a folder called findReference and add the preprocessed images of the dataset with their masks:
- subject1_prep.mha
- subject1_f.mha
- subject2_prep.mha
- subject2_f.mha
- subject3_prep.mha
- subject3_f.mha
File nomenclature has be as follows:
The file name root of image and corresponding mask has to be the same
The image name has to end in
_prep.mhaThe mask name must end in
_f.mha
Create the input file:
[1] ./findReference
[2] r subject2_prep.mha
[3] m subject1_prep.mha
[4] m subject2_prep.mha
[5] m subject3_prep.mha
[6] m etc.
where:
Line 1: findReference folder, containing all the images of the dataset
Line 2: Reference image, indicated as
rLine 3-5: Moving images, indicated as
m
Note that in this example subject2_prep.mha is both the reference and an image of the dataset, because we want to include it as a possible candidate for being the final reference
If you run multiple studies, created the input file for every seed image, adapting the reference (r) file name
Executing findReference.ipynb¶
For each seed image, in findReference.ipynb customize input_file_name and n_of_cores
Launch findReference.ipynb. It will run until convergence or until the number of iterations reaches 10 (If you run the source code, you can change the number of iterations in the file find_reference_for_nb.py, function find_reference, variable maxIterationNo)
Output: Convergence plot¶
The output of the computation is a convergence plot. The graph can reach a plateau or can be zig-zagged. In this last case, choose the reference with the lowest error (y-axis). If the graph shows less than 10 iterations it means that the current reference image is the same as the one in the previous loop.
Segmentation quality¶
You can quantify segmentation quality when a ground truth segmentation is present, whereas you can evaluate segmentation quality only visually when a ground truth segmentation is not available
The metrics we use to evaluate segmentation quality are:
Measures of overlap agreement: Dice coefficient, Jaccard coefficient, and volume similarity, which quantify the overlap between ground truth segmentations and pyKNEEr segmentations
Measure of surface distance: Average of the Euclidean distances between ground truth segmentations and pyKNEEr segmentations
The Jupyter notebook to evaluate segmentation is segmentation_quality.ipynb
Input: Image list¶
Create the input file:
[1] ./segmented
[2] ./segmented_groundTruth
[3] s subject1_prep_fc.mha
[4] g subject1_groundTruth_fc.mha
[5] s subject2_prep_fc.mha
[6] g subject2_groundTruth_fc.mha
[7] m etc.
where:
Line 1: Segmented folder, containing the masks obtained with pyKNEEr
Line 2: Ground truth folder, containing ground truth masks
Lines 3,5: Segmentations obtained with pyKNEEr, indicated as
sLines 4,6: Ground truth segmentations, indicated as
g
Execution, Output, and Visualization¶
Execution:
In
segmentation_quality.ipynb, customizeinput_file_name,output_file_name_overlap, andoutput_file_name_distances, and execute
Output and visualization:
Results will be visualized as graphs and tables, and will be saved in the
.csvfiles for possible subsequent analysis
References¶
[1] Klein S., Staring M., Murphy K., Viergever M.A., Pluim J.P.W. elastix: A Toolbox for Intensity-Based Medical Image Registration. IEEE Transactions on Medical Imaging. vol. 29, no. 1, pp. 196 - 205, January. 2010.
