Introduction¶
pyKNEEr is an image analysis workflow for open and reproducible research on femoral knee cartilage
Characteristics of pyKNEEr are exensively covered in our paper. Find below a concise summary.
Workflow¶
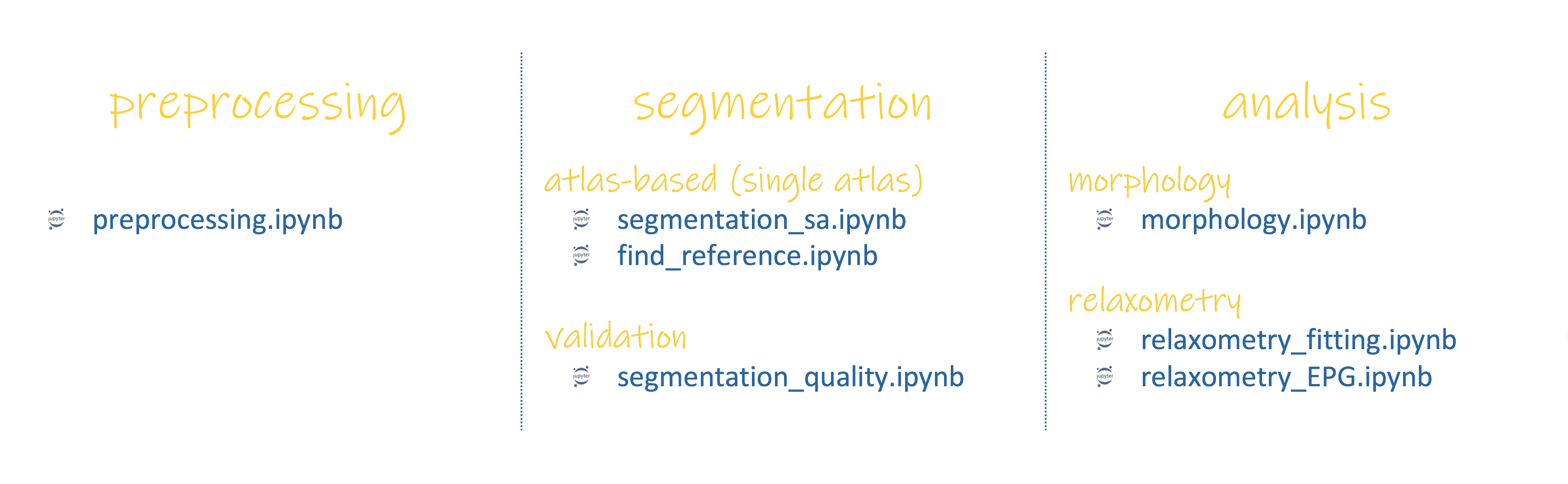
pyKNEEr workflow is composed of three modules:
Preprocessing to homogenize spatial characteristics and enhance intensities
Segmentation of femoral knee cartilage using an atlas-based method
Analysis of cartilage morphometry (thickness and volume) and relaxometry (fitting and EPG modeling)
Code¶
pyKNEEr:
Jupyter Notebooks¶
Jupyter notebooks are the user-interface of pyKNEEr. Once run, you can attached them directly to your paper for computational reproducibility of the results.
For each module of pyKNEEr there are several Jupyter notebooks (*.ipynb), independent from each other:
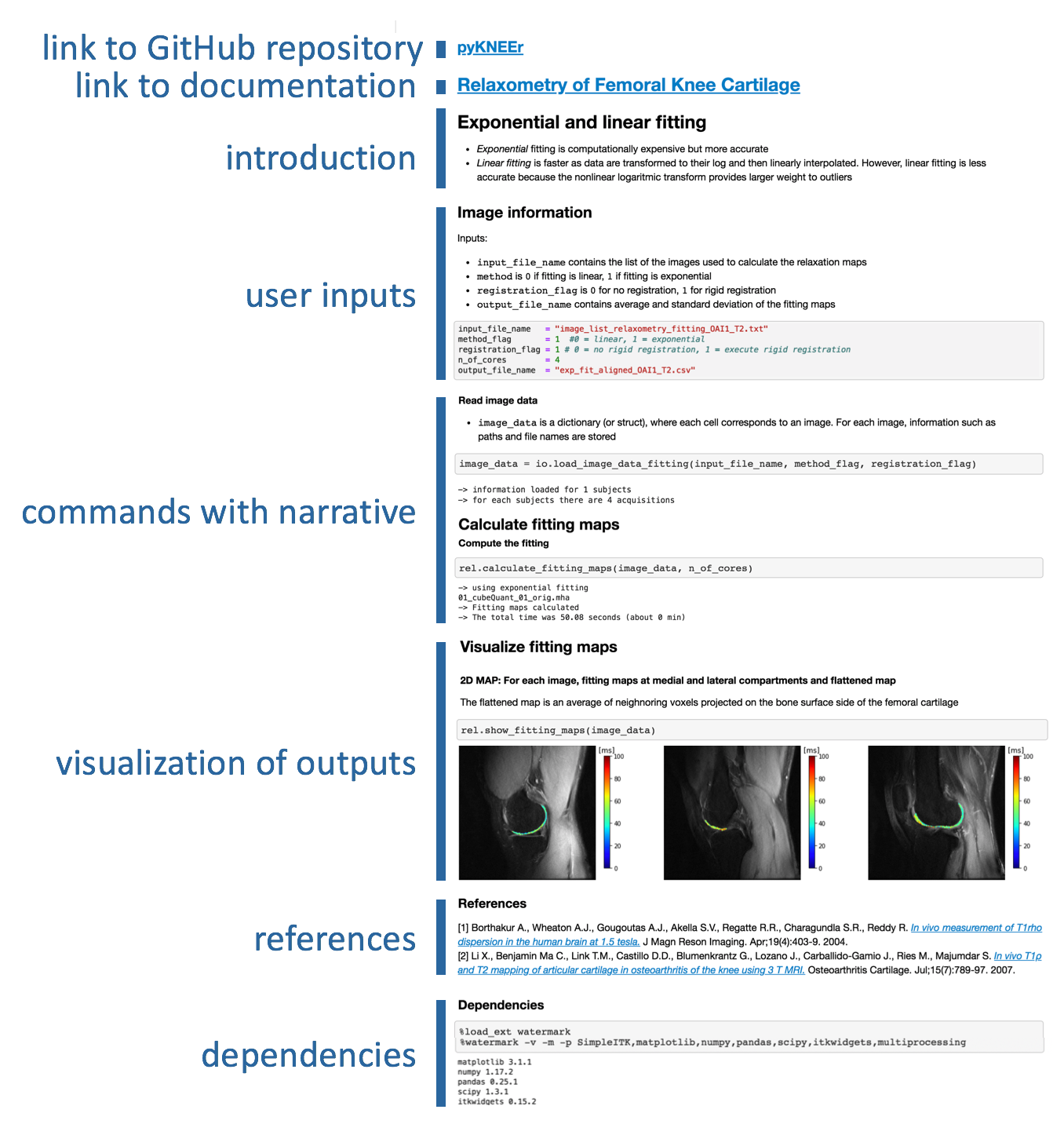
All notebooks have a similar structure:
Data structure¶
pyKNEEr requires data folders to be structured as follows:

Where:
reference: contains the reference (or target) image for the atlas-based segmentation
original: contains dicom folders of the images to analyze
preprocessed: contains images after preprocessing
registered: contains by-products of registration and it can be deleted after computations
segmented: contains segmentation masks
morphology: contains cartilage surfaces, thickness, and volume
relaxometry: contains masked relaxometry maps
Data file formats¶
Inputs and output files are in open formats:
dicom (.dcm): Format of original images to be processed
metafile (.mha): Format of images throughout the workflow
text (.txt): Format of image list inputs and morphology outputs
tabular (.csv): Format of output descriptive statistics in morphology and relaxometry analysis
