Preprocessing¶
Preprocessing is fundamental to provide standardized high quality images to the segmentation algorithm, and thus obtain successful segmentation.
In pyKNEEr preprocessing acts on:
Spatial image characteristics: Images are transformed to the same orientation (RAI, i.e. right, anterior, posterior), knee laterality (left), and origin (0,0,0)
Image intensities: In all images, the homogeneous magnetic field is corrected, intensities are rescaled to the fix range [0,100], and cartilage contours are enhanced [1]
Note
Standardization of spatial characteristics is recommended for all images
Standardization of image intensities is recommended only for images that are going to be segmented
Input: Image folder list¶
For the demo images, the input file is image_list_preprocessing.txt, which contains:
[1] ./original/
[2] 01/DESS/01
[3] right
[4] 01/DESS/02
[5] right
[6] 01/cubeQuant/01
[7] right
[8] 01/cubeQuant/02
[9] right
[10] 01/cubeQuant/03
[11] right
[12] 01/cubeQuant/04
[13] right
where:
Line 1: Path of the folder
original, containing the image dicom foldersEven lines from 2 to 12: Folder names of the dicom stacks to preprocess
Odd lines from 3 to 13: Knee laterality
Tip
When using your own data:
- Create a folder original and add your dicom folders
- Customize image_list_preprocessing.txt with the paths of your own dicom folders
- Specify knee laterality using pyKNEEr coordinate system
- Tip: For images that are not directly involved in intersubject segmentation, you can run a separate notebook where you can set intensity_standardization = 0 to save computational time
Executing preprocessing.ipynb¶
To preprocess data:
Launch Jupyter notebook
In File Browser, navigate to
preprocessing.ipynb, open it, and:Customize the input variables:
n_of_cores(How do I choose the number of cores?)intensity_standardization(0 for only spatial preprocessing, 1 for spatial and intensity preprocessing)
Follow the instructions in the notebook
Save your notebook at the end of the process
Output: Preprocessed images
The preprocessed images are in the folder ./preprocessed. For each dicom folder in the original folder, the outputs are:
*_orig.mha(e.g.01_DESS_01_orig.mha): Images with the same intensities as the volume in the original dicom folder, but different orientation, knee laterality (if right), and image origin. These images can be used to compute relaxation maps*_prep.mha(e.g.01_DESS_01_orig.mha): Images with the same spacial characteristics as*_orig.mhabut different intensities, because the constant magnetic field was corrected, the intensities were rescaled to a fix range, and the cartilage contours were enhanced. These images can be used to segment femoral knee cartilage*_orig.txt(e.g.01_DESS_01_orig.txt): Text files containing the header of the.dcmfiles. They can be used to extract acquisition information such as echo time, flip angle, etc.
Note
Both *_orig.mha and *_prep.mha are anonymized images, while *_orig.txt contains all the information of the dicom header (including subject name, etc.) if the dicom was not anonymized
Visualization: Original and preprocessed images¶
For a qualitative check, for each subject you can see a 2D slice of *_orig.mha and *_prep.mha, similarly to this one:
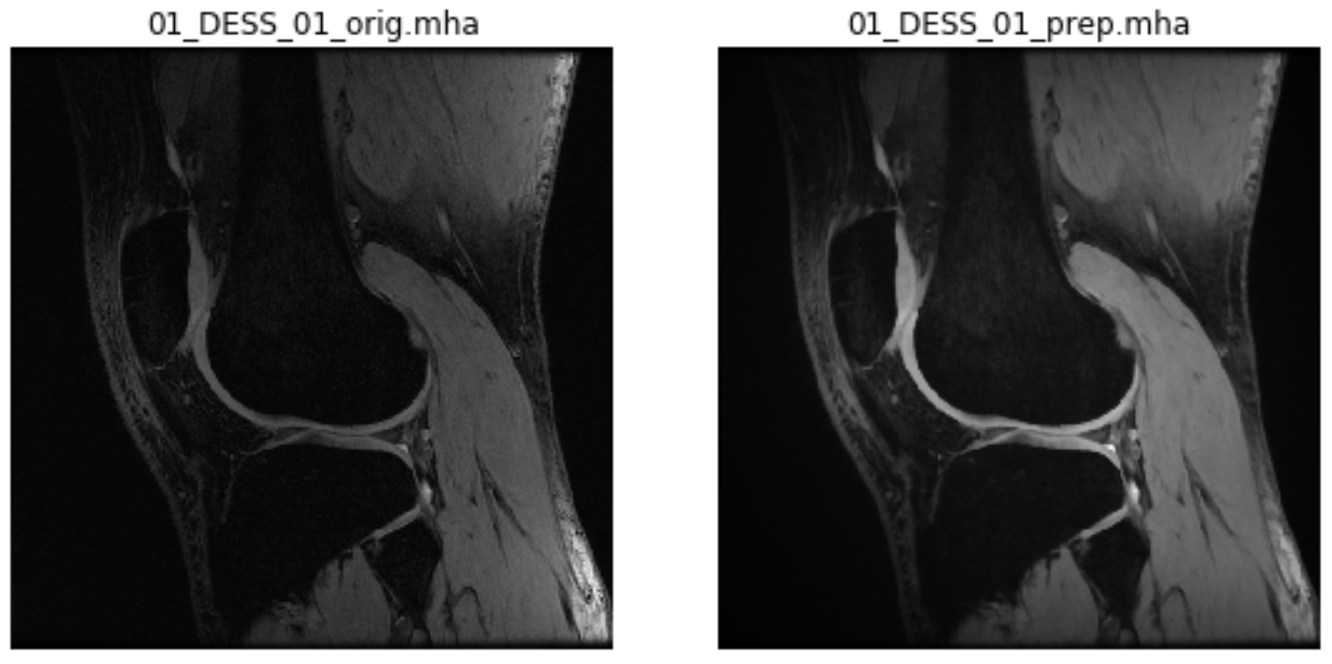
For images that were only spatially standardized, you will see only one 2D slice of *_orig.mha.
For 3D visualization, consider using a medical image software such as ITK-SNAP, which allows comparing images in the same coordinate system
References¶
[1] Shan L., Zach C., Charles C., Niethammer M. Automatic Atlas-Based Three-Label Cartilage Segmentation from MR Knee Images. Med Image Anal. Oct;18(7):1233-46. 2014.
